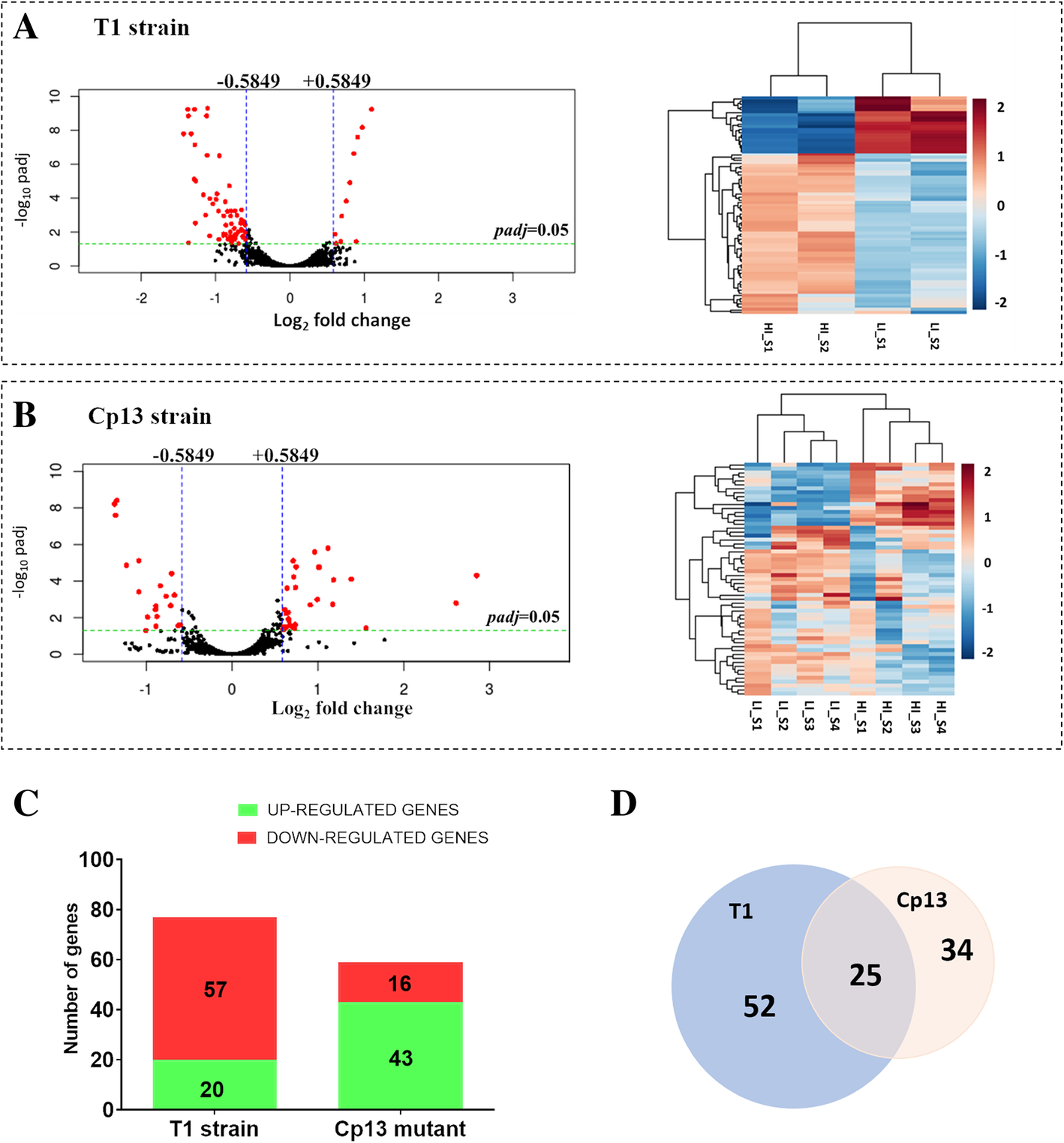
Transcriptome profile of Corynebacterium pseudotuberculosis in response to iron limitation | BMC Genomics | Full Text

Impact of the spotted microarray preprocessing method on fold-change compression and variance stability. - Abstract - Europe PMC
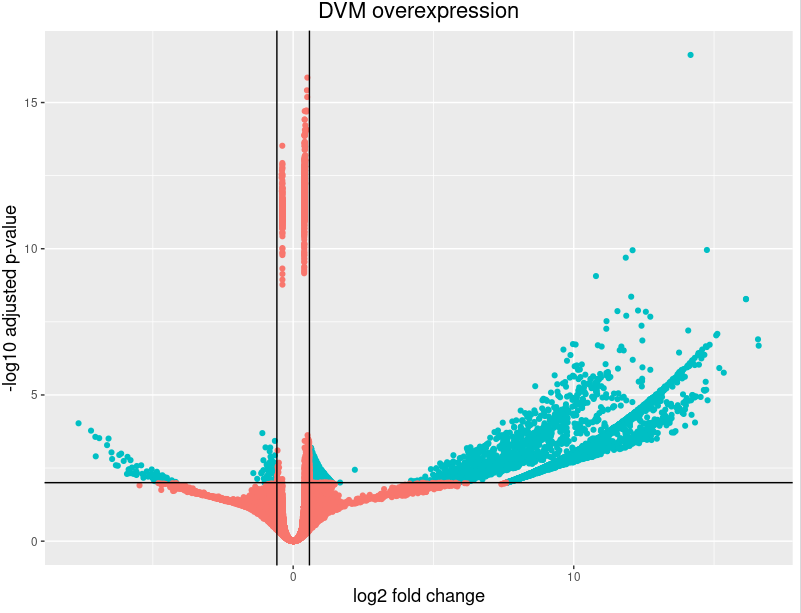
Volcano plot showing log2 (fold change, FC) against −log10 (p-value) of... | Download Scientific Diagram

Volcano plot shows the gene statistical significance (on the y-axis) over the log2 fold change after normal shrinkage (on the x-axis) for all the samples from differential expression analysis between anti-miR-71-2´OMe vs

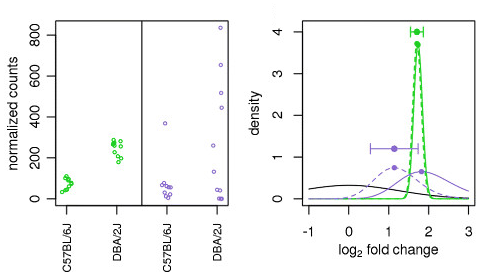
The correlation between log2 (fold change) of circRNAs and log2 (fold... | Download Scientific Diagram

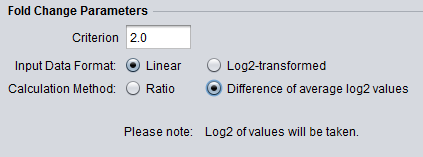
Different logFC (log2foldchange) values for genes from limma-voom and other tools (edgeR and DESeq2)